CRISPR/Cas – genome editing is becoming increasingly popular
The number of publications and patents that involve the CRISPR/Cas system has been increasing exponentially since the technique was first described a few years ago. The increase in funding for projects involving CRISPR/Cas also demonstrates how powerful this new method is. The targeted modification of genomes (also called gene editing or genome editing) using CRISPR/Cas is extraordinarily accurate and also has the potential to cure hereditary diseases. However, the benefits of genome editing raise ethical concerns and involve risks that need to be taken seriously.
When researchers applied CRISPR/Cas for the first time in 2012, nobody could foresee the impact the method would have on science. Particularly as the researchers involved in the project were not at all focused on testing the suitability of CRISPR as a genome-engineering tool. However, the potential of CRISPR/Cas soon became clear. It is generally assumed that the system with the complex name “clustered regularly interspaced short palindromic repeats” (CRISPR)/”CRISPR-associated” (Cas), is the next major scientific revolution after the development of the polymerase chain reaction (PCR). Experts believe that the developers of the method are likely to be awarded the Nobel Prize in Chemistry at some stage in the future. CRISPR/Cas is applied in many research areas where genes and genomes need editing, i.e. modifying. The optimisation of the method has led to extraordinary progress in the field of genome editing.
CRISPRs have been known about for quite some time. Clustered repeats were first described in E. coli in the 1980s and more recently have been shown to provide bacteria and archaea (prokaryotes) with a form of acquired immunity. It has also been shown that the spacers between CRISPR repeats are homologous to phage DNA sequences. 30 years after the initial discovery of the repeats, researchers around the world are able to explain how these repetitive elements are generated and their work benefits greatly from an in-depth knowledge of CRISPRs.
Cheap, fast and simple
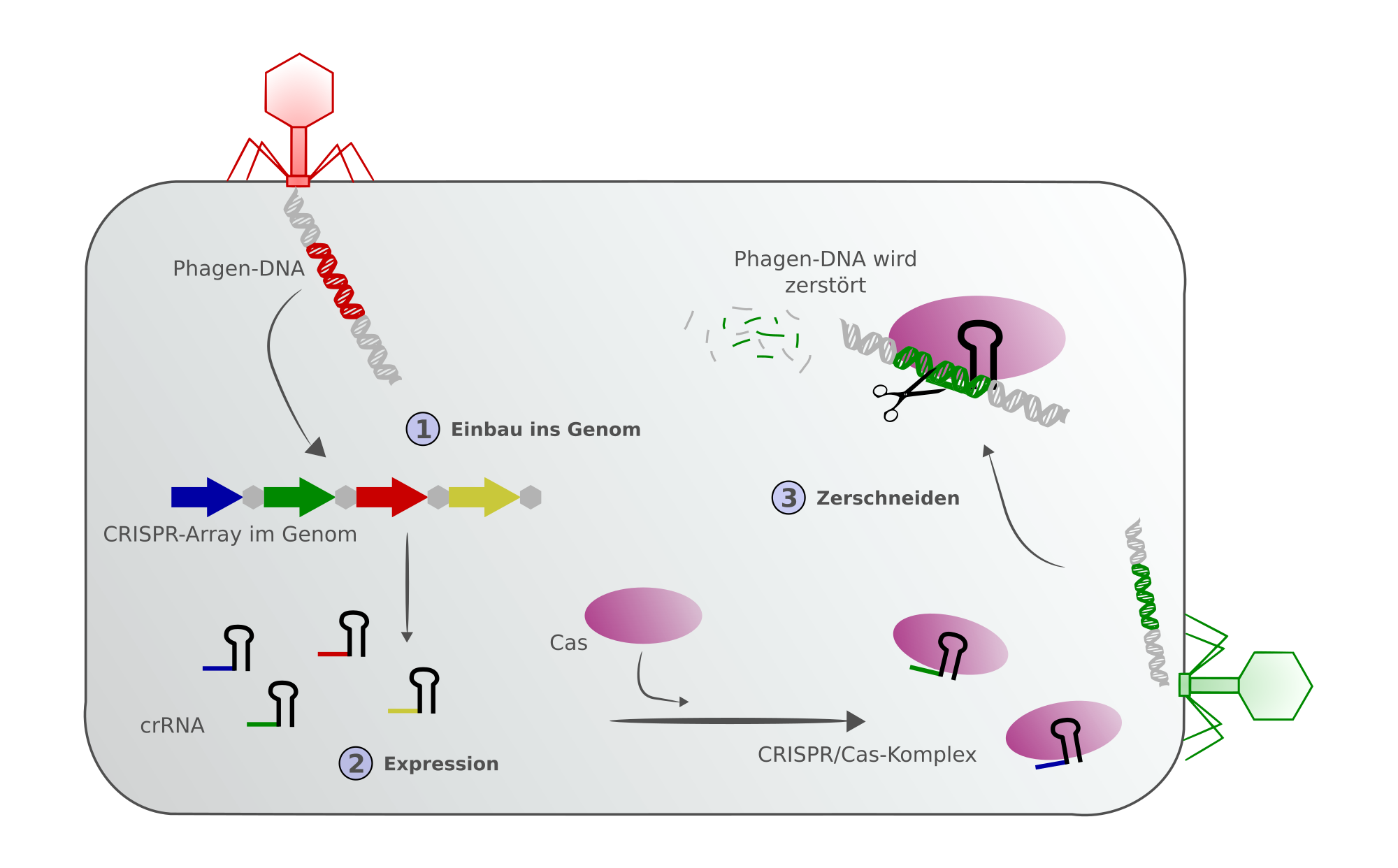
A prokaryote that is infected by a phage for the first time incorporates parts of the phage genome as new spacers in its CRISPR locus; this is part of the immune response to phage infection. The genome fragment is expressed as crRNA (CRISPR-RNA), associated with the Cas protein (a DNA-cutting endonuclease enzyme), and is guided to the target DNA, i.e. the phage genome from which the spacer originated, which is therefore homologous to the crRNA. Cas cuts the appropriate DNA, which is then degraded. This prevents the bacterium from being infected by the phage, making it a similar process to the innate immune system in humans, which when first encountering a pathogen, establishes an immune response specific for the particular pathogen. However, what makes CRISPR/Cas particularly interesting is its suitability for editing genes. The technique has huge potential in a wide variety of applications. crRNA homologous to the target DNA is synthesised in the laboratory and used to guide the CRISPR/Cas9 complex to the correct section of the host DNA where it induces a blunt-end break in the DNA double strand. This also works in eukaryotic cells with much more complex genomes than prokaryotes. The double-strand break is repaired by one of the following repair pathways: the efficient but error-prone non-homologous end joining pathway or the homologous recombination pathway, which is less efficient, but highly accurate. The cell-cycle-dependent homologous recombination pathway is of greater importance for the application of the technique. An artificial DNA repair template which is homologous to the cut DNA fragment guides the cellular repair process and allows the specific mutation of the DNA sequence as well as the insertion or removal of specific DNA sequences. CRISPR can also be used to remove large chromosomal regions or to move them to a different place. It is thus possible to modify any information on the DNA: genes, gene cassettes and important regulatory elements in coding as well as non-coding regions.
The continual improvements in the efficiency and accuracy of the system are of particular interest. Around ten years ago, researchers around the world were excited about the new possibilities of DNA-cutting zinc finger nucleases (ZFN). However, along with the transcription activator-like effector nucleases (TALEN), which had to be assembled from several modules on the protein level, they did not have the desired outcome; they were too expensive, ineffective and difficult to engineer. What scientists really wanted was a tool that could be used to insert and remove defined DNA segments, a process that once took several years, but can now be done more rapidly. All researchers need to do is create the crRNA that guides the Cas endonuclease to the target sequence. The introduction of the CRISPR/Cas complex into eukaryotes does not present any particular difficulties either and can be achieved biochemically (by adding the complex to the cells) or genetically (by introducing the relevant genetic endonuclease blueprint into the cells).
The new and highly precise genome-cutting tool opens up many new possibilities. While it is already widely used in R&D laboratories, it is hoped that it will soon be used for treating diseases. It would greatly improve the prospects for treating hereditary diseases such as cystic fibrosis and haemophilia. CRISPR/Cas may also be of help in treating genome mutations characterised by the lack of an entire chromosome or the presence of two chromosome copies instead of just one (known as aneuploidies such as trisomy 21). As CRISPR/Cas works in principle in any organism whatsoever, there is no reason why this should not be possible.
A genome-edited plant might not necessarily be a GMO

While the new technologies clearly offer new opportunities, there are also risks involved. Most research projects are devoted to pure research, for example investigating gene function. However, in the medium term, the technology is expected to be increasingly used in plant and animal breeding. The clarification of ethical issues and the formulation of regulations will need to keep pace with the rapid progress of the new genome-editing tools.
Plants that are modified using CRISPR/Cas in order to make them more resistant, for example by inserting small DNA sequences or exchanging a single nucleotide, cannot be differentiated from their naturally occurring counterparts that have acquired these properties by way of spontaneous mutations or breeding. Paragraph 3 of the German Genetic Engineering Act (GenTG) stipulates: “A genetically modified organism (GMO) is an organism, with the exception of human beings, in which the genetic material has been altered in a way that does not occur naturally by mating or natural recombination.” According to this definition, CRISP/Cas-edited organisms should not be considered GMOs. However, this statement can currently only be considered provisional. The problem is that it is impossible to find out whether the DNA of an organism has been modified using CRISPR/Cas or whether the modification occurred naturally. There are simply no biological or chemical markers that can detect this change. Providers of genome-edited crops hope that this will make it easier to eventually bring gene-edited plants to market, even genetically intricate polyploids such as wheat. It will be interesting to see whether the European Union calls for the regulation of CRISPR/Cas-edited crops or not.
Prof. Dr. Detlef Weigel from the Max Planck Institute for Developmental Biology in Tübingen and other researchers from Germany, the USA and China proposed a regulatory framework for genome-edited crops. They suggested the following as key guiding principles when considering the generation and regulation of genome-edited crops: the exact documentation of DNA modifications and the removal of all foreign sequences, minimising the risk of escape of genome-edited crops from fields during the research and development phase, and identifying the phylogenetic relationship of the edited plant and the gene donor if new sequences are introduced. Weigel suggests that plants should be seen as the result of conventional breeding wherever it is impossible to demonstrate whether or not they were generated by genetic engineering.
Power that needs to be used wisely
The use of genome-editing techniques for manipulating animal DNA is more complicated and controversial. That said, we need to bear in mind that the genetic manipulation of animals is not a contemporary invention. The first attempts were made around thirty years ago, but problems such as disease, infertility and inefficiency dampened the initial euphoria. The old methods used to manipulate DNA did not lead to satisfactory results. Genetically modified salmon, which the American Food and Drug Administration (FDA) consider as safe to eat as non-genetically engineered salmon is, to date, the only genetically modified animal or animal product that has been approved for human consumption. The original idea was to create robust livestock that grows quickly, is able to consume phosphate-rich food or produce lactose-free milk. In recent years, work on the genetic modification of animals has been mainly based on the hope of producing medically relevant compounds or using the animals as organ donors. The new genome-editing methods have given new impetus to these fields.
Genetic engineering was first used for treating human diseases in the 1990s. It was finally possible to beat hereditary diseases – at least in cell cultures and albeit with a large number of adverse effects. As expected, CRISPR/Cas turned out to be highly suitable for modifying human DNA, resulting in a conflictual debate about the ethical aspects of genome editing in humans. The feared, though expected piece of news was not long in coming: in April 2015, Chinese researchers reported that they used CRISPR/Cas to modify human embryos. Genetic changes to early embryos are known as germline modifications and the effect of such embryos on future generations remains unpredictable. The Chinese publication re-ignited the ethical debate and there are growing concerns about unethical uses and risks of the technique. Opponents claimed that the modifications introduced (including unintended off-target mutations at other DNA sites) might be passed on to the next generation and that the simplicity of the CRISPR/Cas system enables many researchers to master the technical hurdles of human genome editing. The IAG (Interdisciplinary Gene Technology work group) argues in favour of a moratorium on germline experiments involving humans while the potential benefits and risks of the technology are assessed. The results of a second study introducing precise genetic modifications into human embryos using CRISPR/Cas editing were published in April 2016.
Its simplicity, versatility and high level of efficiency have made CRISPR/Cas a very successful technique. While in evolutionary terms, it only cleaves and destroys DNA, scientists have turned the system into a practical tool with unprecedented precision. The coming years will show exactly what CRISPR/Cas can be used for. In the field of molecular biology, a new era has already begun. Maybe we would all be well advised to spend more time evaluating this double-edged development.
Further reading
1 Huang S, Weigel D, Beachy RN, Li J. A proposed regulatory framework for genome-edited crops. Nat Genet. 2016 Jan 27;48(2):109-11. doi: 10.1038/ng.3484.
2 Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015 May;6(5):363-72. doi: 10.1007/s13238-015-0153-5.
3 Jarasch ED: "Call for a moratorium on germ line experiments in humans"
4 Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing .J Assist Reprod Genet. 2016 May;33(5):581-8. doi: 10.1007/s10815-016-0710-8.